Research Areas and Projects
Identification of candidate anti-cancer molecules
Recent WHO-IARC data reports that cancer is the major cause of morbidity and mortality, with approximately 14 million new cases and 8 million cancer-related deaths in 2012 (GLOBOCAN-2014). Traditional chemotherapeutic agents remain inefficient for the treatment of cancer with poor prognosis. Therefore there is a need for targeted therapies. Our group has been working on the molecular biology of liver cancer with a specific aim of discovery of novel candidate molecules for the treatment of this deadly Disease. Our focus is liver cancer, which is the 5th most common and 3rd lethal cancer and associated with poor overall survival. Major molecular and cellular biology approaches
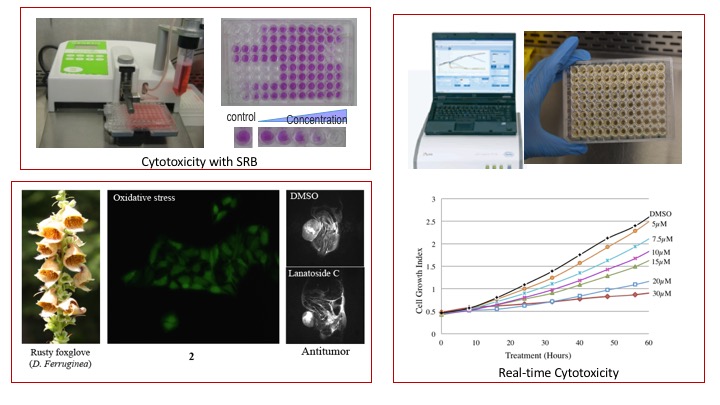
- NCI-60 Cytotoxicity screening of small molecules on liver, colon and breast cancer cells
- Evaluation of cytotoxicities in real time with RT-CES systems
- Investigation of the induced morphological changes
- Cell cycle experiments
- Wound healing and migration assay with time lapse camera
- Characterization of cell death mechanism
- Mice xenograft experiments